Instructor
 |
Department of Genetics, Cell Biology and Anatomy 402-559-5925 |
Education:
PhD, University of Mysore, Karnataka, India, 2013
Post-doc, NYU School of Medicine, New York, NY, 2020
Research:
My research is centered around integrating genetics and computational genomics, with a focus on developing novel tools and pipelines to model biological systems and investigate the complexities of human diseases and disorders.
|
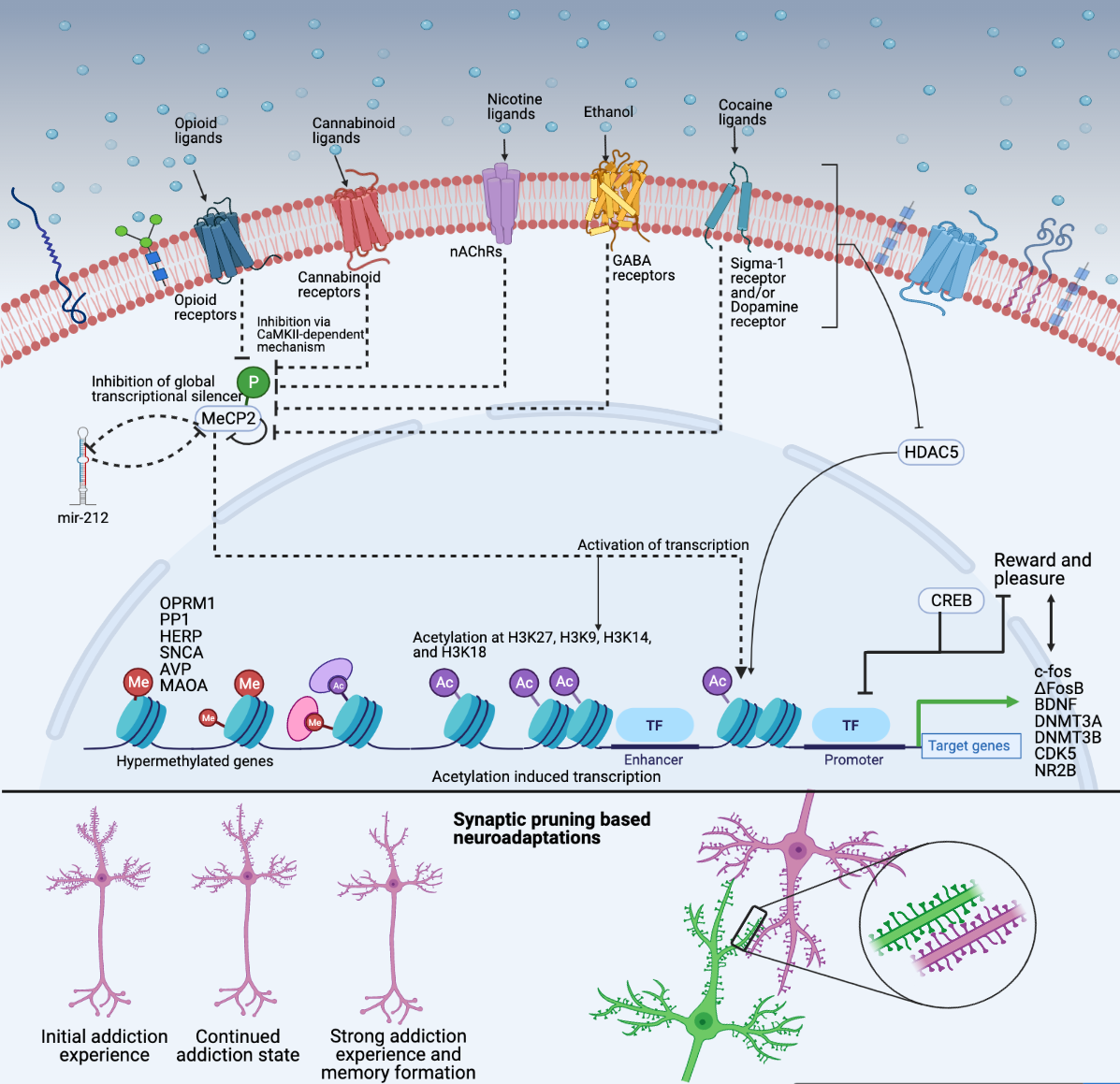
Substance use circuitry. Substance use disorder (SUD) is a complex and multifaceted condition characterized by persistent craving, pleasure, and reward that ultimately leads to addiction. My research focuses on the molecular and neural mechanisms underlying the development and progression of SUD, as well as the de-addiction process. Specifically, I am interested in understanding how chronic substance use induces maladaptive circuits in the brain that lead to addiction. I hypothesize that chronic substance use induces significant alterations in the global transcriptome signatures of the brain, leading to pruning of maladaptive neural circuits that drive addiction. Furthermore, I believe that comparing global transcriptome signatures between chronic and acute substance users, as well as those in withdrawal, will help in understanding the various waypoints in the evolving landscape of substance user brains. By gaining a better understanding of the molecular and neural mechanisms involved in addiction, we can develop more effective treatment strategies to combat SUD. However, recognizing the neural circuits related to substance addiction requires a synthesis of OMICS perspectives to delineate how cell-autonomous molecular adaptations in vulnerable individuals drive the altered functioning of the larger neural circuits within which those cells operate, and how circuit-level disruptions caused by drugs contribute to the cells' molecular adaptations. We explore DNA variants that impart susceptibility and use transcriptome and chromatin state assays to further understand the dynamics guided by footprinting, transcription factor co-occurrence, motif enrichment, transcriptional networks, and circDNA to best answer through an iterative process. |
|
Publications:
- AM Veerappa, P Padakannaya and NB Ramachandra. Copy number variation-based polymorphism in a new pseudoautosomal region 3 (PAR3) of a human X-chromosome- transposed region (XTR) in the Y chromosome. Functional & Integrative Genomics, Springer, 2013, 1-9.
- AM Veerappa, M Saldanha, P Padakannaya and NB Ramachandra. Family-based genome- wide copy number scan identifies five new genes of dyslexia involved in dendritic spinal plasticity. Journal of Human Genetics, Nature Publishing Group, 2013, 1-8.
- AM Veerappa, S Vishweswaraiah, K Lingaiah, M Murthy, DS Manjegowda, R Nayaka and NB Ramachandra. Unraveling the Complexity of Human Olfactory Receptor Repertoire by Copy Number Analysis across Population Using High Resolution Arrays. PloS one, Public Library of Science, 2013, e6684.
- AM Veerappa, M Saldanha, P Padakannaya and NB Ramachandra. Genome-wide copy number scan identifies disruption of PCDH11X in developmental dyslexia. American Journal of Medical Genetics, Part B: Psychiatry Genetics, 2014, 162 (8) 889-897.
- AM Veerappa, M Saldanha, P Padakannaya and NB Ramachandra. Family based genome- wide copy number scan identifies complex rearrangements at 17q21.31 in dyslexics. American Journal of Medical Genetics, Part B: Psychiatry Genetics. 2014,165 (7); 572-580.
- S Vishweswaraiah, AM Veerappa, PA Mahesh, BS Jayaraju, SK Chaya, and NB Ramachandra. Molecular Interaction Network and Pathway Studies of ADAM33 Potentially Relevant to Asthma. Annals of Allergy, Asthma & Immunology, 2014, 133 (4): 418-424.
- S Vishweswaraiah, AM Veerappa, PA Mahesh, and NB Ramachandra. Copy Number Variation burden on Asthma sub-genome in normal cohorts identifies susceptibility Allergy, Asthma & Immunology Research, 2014, 7(3): 265-75.
- AM Veerappa, S Vishweswaraiah, K Lingaiah, MN Murthy, RV Suresh, DS Manjegowda, and NB Ramachandra. Global spectrum of Copy Number Variations reveals genome organizational plasticity and proposes new migration routes. PloS ONE, Public Library of Science, 2015, 10(4): e0121846.
- MN Murthy, AM Veerappa, K Belur, and NB Ramachandra. High resolution arrays reveal burden of Copy Number Variations on Parkinson Disease genes associated with increased disease risk in random cohorts. Neurological Research. 2016 Sep;38(9):775-85.
- T Prakash, AM Veerappa, and NB Ramachandra. Complex interaction between HNRNPD mutations and risk polymorphisms is associated with discordant Crohn’s disease in monozygotic twins. Autoimmunity. 2017 Aug;50(5):275-276.
- Y Kattimani and AM Veerappa. Dysregulation of NRXN1 by mutant MIR8485 leads to calcium overload in pre-synapses inducing neurodegeneration in Multiple Sclerosis. Multiple Sclerosis and Related Disorders. (2018). 22:153-156.
- U Radhakrishna, S Vishweswaraiah, AM Veerappa, R Zafra, S Albayrak, PH Sitharam, NM Saiyed, NK Mishra, C Guda, R Bahado-Singh. Newborn blood DNA epigenetic variations and signaling pathway genes associated with Tetralogy of Fallot (TOF). PLoS One. (2018) Sep 13;13(9) :e0203893.
- Y Kattimani and AM Veerappa. Complex interaction between mutant HNRNPA1 and gE of Varicella Zoster Virus in pathogenesis of Multiple Sclerosis. (2018). Autoimmunity. 2018 Jun; 51(4):147-151.
- Veerappa AM. Cascade of interactions between candidate genes reveals convergent mechanisms in keratoconus disease pathogenesis. Ophthalmic Genet. 2021 Apr;42(2):114- 131. doi: 10.1080/13816810.2020.1868013. Epub 2021 Feb 8. PMID: 33554698.
- Veerappa AM, Pendyala G, Guda C. A systems omics-based approach to decode substance use disorders and neuroadaptations. Neurosci Biobehav Rev. 2021 Nov; 130:61-80. doi: 10.1016/j.neubiorev.2021.08.016. Epub 2021 Aug 17. PMID: 34411560; PMCID: