Assistant Professor
 |
Department of Genetics, Cell Biology and Anatomy 402-836-9794 |
Education:
BS, Brigham Young University, Provo, UT, 2002 - 2008
PhD, University of Michigan, Ann Arbor, MI, 2009 - 2014
Emory University, Atlanta, GA, 2015 - 2019
Honors and Awards:
K99/R00 Pathway to Independence Award, NIGMS, 2018 - 2022
|
Research: |
|
|
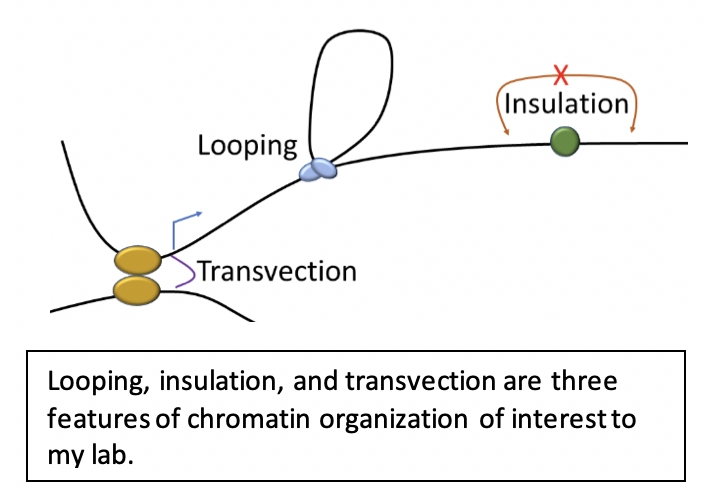
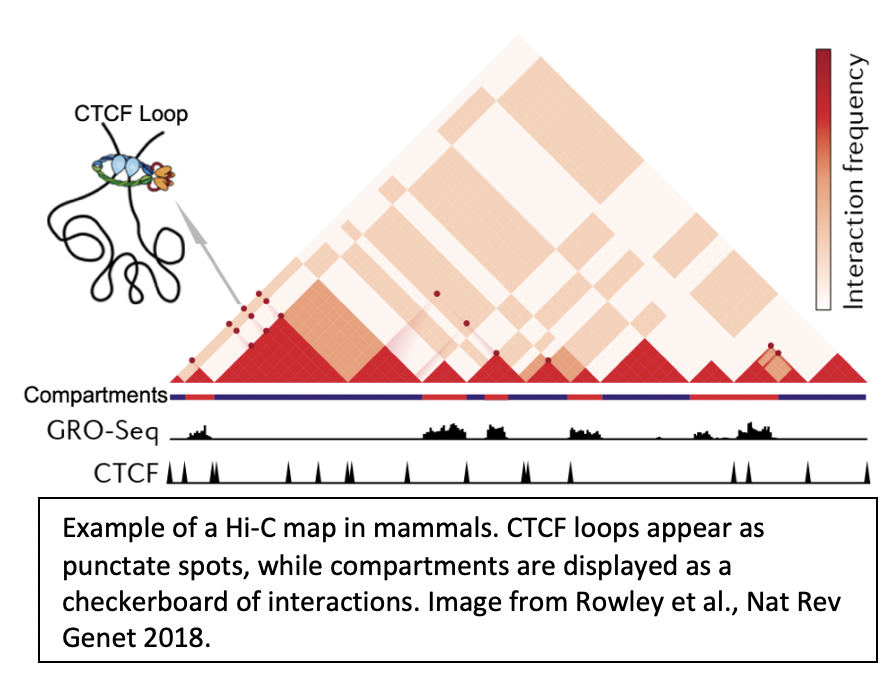
1. How are chromatin loops formed? |
 |
|
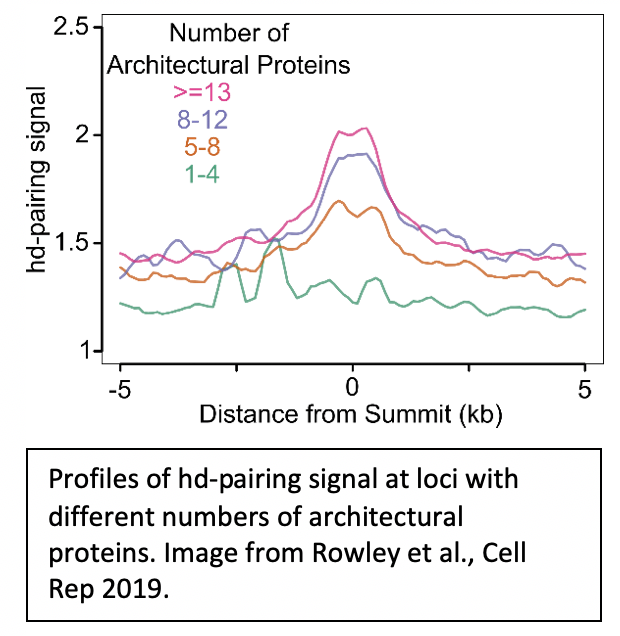
2. How do architectural proteins contribute to pairing and transvection? |
|
|
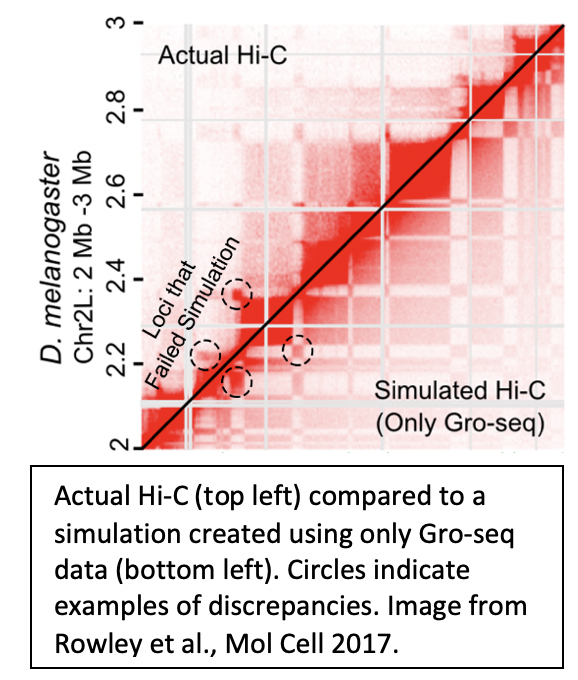
3. Predicting 3D chromatin organization |
 |
Publications:
- Cummings CT, Rowley MJ. Implications of Dosage Deficiencies in CTCF and Cohesin on Genome Organization, Gene Expression, and Human Neurodevelopment. Genes, 2022. PMID: 35456389.
- Rocks D, Shukla M, Ouldibbat L, Finneman SC, Kalluchi A, Rowley MJ, Kundakovic M. Sex-specific multi-level 3D genome dynamics in the mouse brain. Nature Communications. 2022 PMID: 35705546.
- Mirza S, Saleem I, Kalluchi A, Raza M, Pal D, Mohapatra B, Lele S, Qiu F, Yu L, Zheng Z, Zhang Y, Alsaleem MA, Rakha EA, Band H, Rowley MJ, Band V. Ecdysoneless protein regulates viral and cellular mRNA splicing to promote cervical oncogenesis. Mol Cancer Res 2022 (co-corresponding author). PMID 34670863
- Engstrom AK, Walker AC, Moudgal RA, Myrick DA, Kyle SM, Bai Y, Rowley MJ, Katz DJ. The inhibition of LSD1 via sequestration contributes to tau-mediated neurodegeneration. PNAS 2020. PMID 33139560.
- Rowley MJ, Poulet A, Nichols MH, Bixler BJ, Sanborn AL, Brouhard EA, Hermetz K, Linsenbaum H, Csankovszki G, Lieberman Aiden E, Coreces VG. Analysis of Hi-C data using SIP effectively identifies loops in organisms from C. elegans to mammals. Genome Research. 2020 PMID: 32127418.
- Gutierrez-Perez I, Rowley MJ, Lyu X, Valadez-Graham V, Vallejo DM, Ballesta-Illan E, Lopez-Atalaya JP, Kremsky I, Caparros E, Corces VG, Dominguez M. Ecdysone-Induced 3D Chromatin Reorganization Involves Active Enhancers Bound by Pipsqueak and Polycomb. Cell Rep. 2019 PMID: 31484080.
- Rowley MJ, Lyu X, Rana V, Ando-Kuri M, Karns R, Bosco G, Corces VG. Condensin II counters cohesin and RNA Polymerase II in the establishment of 3D chromatin organization. Cell Reports. 2019; PubMed PMID: 30865881
- Jung YH, Kremsky I, Gold HB, Rowley MJ, Punyawai K, Buonanotte A, Lyu X, Bixler BJ, Chan AWS, Corces VG. Maintenance of CTCF– and Transcription Factor-Mediated Interactions from the Gametes to the Early Mouse Embryo. Mol Cell. 2019; PubMed PMID: 31056445
- Rowley MJ, Corces VG. Organizational principles of 3D genome architecture. Nat Rev Genet. 2018; PubMed PMID: 30367165
- Lyu X, Rowley MJ, Corces VG. Architectural proteins and pluripotency factors cooperate to orchestrate the transcriptional response to hESCs to temperature stress. Mol Cell. 2018; PubMed PMID: 30122536.
- Rowley MJ, Nichols M, Lyu X, Ando-Kuri M, Rivera S, Hermetz K, Wang P, Ruan Y, Corces VG. Evolutionarily conserved principles predict 3D chromatin organization. Mol Cell. 2017; PubMed PMID: 28826674. Cover article.
- Rowley MJ, Rothi MH, Böhmdorfer G, Kucinski J, Wierzbicki AT. Long-range control of gene expression via RNA-directed DNA methylation. PLoS Genet. 2017; PubMed PMID: 28475589
- Cubenas-Potts C, Rowey MJ, Lyu X, Li G, Lei EP, et al. Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res. 2016; PubMed PMID: 27899590
- Rowley MJ, Corces VG. The three-dimensional genome: principles and roles of long-distance interactions. Curr Opin Cell Biol. 2016; Review. PubMed PMID: 26852111
- Rowley MJ, Böhmdorfer G, Wierzbicki AT. Analysis of long non-coding RNAs produced by a specialized RNA polymerase in Arabidopsis thaliana. Methods. 2013; PubMed PMID: 23707621