Projects
Host Trafficking to the Chlamydial Inclusion
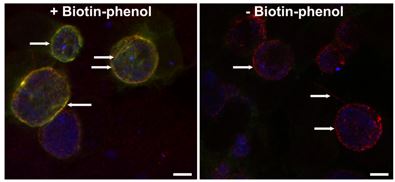
Ctr L2 expressing IncAtm-APEX2, which biotinylates the inclusion membrane in the presence of biotin-phenol. Biotinylated proteins can then be extracted from cell lysates and identified by mass spectrometry.
The laboratory of Lisa Rucks, PhD, focuses on the function of eukaryotic SNAREs (N-ethylmaleimide sensitive attachment protein receptor) at the chlamydial inclusion.
SNARE proteins serve to decrease the energy required to fuse a host vesicle with a target membrane. In this case, the target membrane is the chlamydial inclusion.
We have demonstrated that syntaxins 6 and 10 and VAMPs 3 and 4 localize to chlamydial inclusion. Further, we have demonstrated that syntaxin 6 and VAMP4 (2 out of 4 required proteins to form a fusogenic SNARE complex) interact at the chlamydial inclusion.
We hypothesize that the chlamydial inclusion specifically intercepts multiple fusogenic SNARE complexes to create and maintain a defined lipid composition in the inclusion membrane. To study this hypothesis, we use proximity labeling strategies to identify membrane-protein protein interactors.
We are also examining the impact of the loss of these specific SNARE proteins on chlamydial growth, development, and organismal membrane organization. We are also developing high resolution imaging techniques to try to visually capture these biochemical interactions.
How Chlamydial Membranes Are Organized
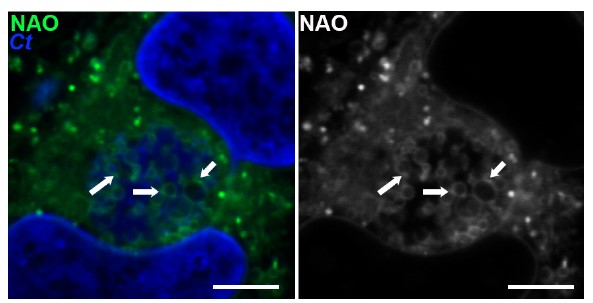
Chlamydia trachomatis (Ct, blue) are stained with a neutral lipid stain. Arrows indicate areas of lipid concentration.
It has been recently established that bacteria also have organized membranous structures with strikingly similar features to eukaryotic detergent-resistant membranes. In bacteria, the purpose of these rafts is to organize bacterial secretion apparati, kinase signaling networks, and cell division machinery.
Three types of lipids are found within both eukaryotic and prokaryotic detergent-resistant membrane microdomains: polycyclic terpenoids (cholesterol or hapanoids), noncyclic terpenoids (squalene or carotenoids) and sphingolipids (sphingomyelin and/or cardiolipin).
Chlamydia are rare amongst bacteria in that they incorporate sphingomyelin into their cellular membranes. Additionally, with their reduced genome, they do not have the genetic capacity to support multiple kinase signaling networks, yet they maintain the ability to respond to nutrient stress. Here, we seek to understand how chlamydial membranes are organized, and how altering chlamydial lipid content (by altering eukaryotic pathways that deliver lipids to chlamydia) may impact chlamydial membrane organization by assessing potential changes in to protein content and membrane fluidity of the chlamydial outer membrane.
Consistent with our model, alterations in chlamydial lipid content will alter the organization of chlamydial membranes. We will first examine how chlamydial membranes are organized under normal growth conditions, and characterize visually and biochemically how they are organized under altered lipid acquisition conditions.
How Stress Impacts Chlamydia
Chlamydia have reduced genomes, and lack many of the common bacterial signaling mechanisms that dictate how the organisms respond to stress. As obligate intracellular pathogens, their exposure to extreme environments is limited, but they can be and are subject to nutrient limitation.
Growth in an environment lacking tryptophan results in a state of quiescent viability known as persistence. We have found in our study of SNARE proteins, that a phenotype of a loss of a SNARE can be slowed growth, which begs the question: what are the molecular signals that dictate whether Chlamydia slow their growth versus enter persistence? And during persistence what are the minimal needs (nutrient/carbon sources, host-pathogen interactions to support the chlamydial inclusion) that determine quiescent viability versus death?
This is a collaborative project with Drs. Scot Ouellette and Rey Carabeo of the Obligate Intracellular Research Group.