Brinkworth Laboratory
Chlamydia Evasiveness
Amanda Brinkworth, PhD
Assistant Professor, UNMC Department of Pathology, Microbiology and Immunology

This lab is using innovative advances in tissue engineering, single-cell transcriptomics, and confocal microscopy to reveal the mechanisms used by bacterium to colonize human epithelial tissues and evade initial immune responses at the primary infection site.
More information on Dr Brinkworth's research on vectors themselves can be found here: Brinkworth Lab Vector Research
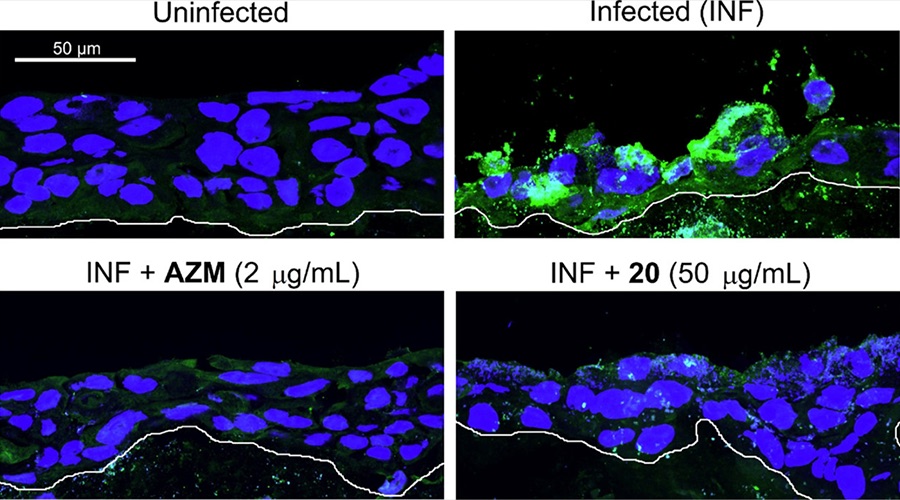
Fluorescent microscopy images of 3D-organotypic ectocervical stratified epithelium infected with Chlamydia trachomatis (green). Treatment with compound 20, a ClpP inhibitor, abolishes infection similar to treatment with Azithromycin.
Chlamydia infection establishment and spread
Despite widespread STD testing and treatment availability, chlamydial genital infections continue to rise annually worldwide, resulting in high medical costs associated with infertility, ectopic pregnancy and pelvic inflammatory disease in women. This problem is exacerbated by the fact that previous chlamydial infection only confers partial immunity at best.
By studying the evasion mechanisms employed during early infection of the female reproductive tract by chlamydia trachomatis, we expect that the immune cell priming defects underlying incomplete immunity will be revealed. Chlamydia trachomatis’s ability to colonize epithelium relies on multiple strategies to prevent cell autonomous immunity and to dampen of cytokine responses in the local microenvironment.
In Fontanilla et al. (MBio 2024. doi: 10.1128/mbio.01834-24 ), we have demonstrated that chlamydia reduces expression of key factors in interferon signaling, including JAKs, STATs, and antimicrobial effectors in infected epithelial cells, which we expect to have a lasting effect on bystander epithelia and immune cells. We utilize co-culture approaches, single-cell RNA-sequencing, and staggered infections to determine the effects of epithelial conditioning by chlamydia on host cell responses and infection spread.
We have leveraged an organotypic in vitro stratified epithelium model to study chlamydial growth and survival in the ectocervix. In fact, Nogueira et al. (Front Cell Infect Microbiol. 2017. doi: 10.3389/fcimb.2017.00438.) revealed that infection of the apical surface of fully stratified epithelium resulted in low numbers of intracellular chlamydia that did not complete development. In collaboration with the Ouellette lab, we have used this model for monitoring antibacterial clearance of the pathogen in the ectocervix (Saleem et al, ACS Infect Dis. 2022 ), and in collaboration with the Carabeo lab, we are using single-cell transcriptomic, proteomic and metabolic approaches to characterize differences between basal, central, and apical cells of the stratified epithelium that may underlie infection susceptibility.
By revealing which nutrient acquisition factors and metabolic enzymes are absent from cells lining the apical surface, we can anticipate alternate metabolic factors chlamydia will need to scavenge from dying cells or nearby microflora to survive. We will use targeted-metabolic labeling approaches to determine which metabolites can be acquired and utilized by chlamydia and define the metabolic requirement for restoration of chlamydial development in the upper layers of the stratified epithelium.
Taken together these approaches will define the metabolic requirements for ectocervical colonization by C. trachomatis and indicate the ectocervix as a reservoir for chronic recurring infections.