Variants of the virus that causes COVID-19 continue to plague the world with spikes in infection, keeping the current pandemic from being fully controlled as Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) infections remain unmanageable in some parts of the world.
Researchers at Bond Life Sciences Center (LSC) and the UNMC recently released a paper documenting their research on mutations in four new variants — B.1.1.7, B.1.351, P.1 and CAL.20C. These evolving variants appear to be more infectious and transmissible than the original Wuhan-Hu-1 virus.
It started with a new spike in infections of a variant of SARS-CoV-2 in the UK, despite a slowdown in infections due to precautions of masks and lockdowns. The new spike meant something about the virus had changed.
“Our goal was to find out where these variants are distributed, and how they are migrating,” said Kamal Singh, assistant director, Molecular Interactions Core, at Bond LSC. Singh was a co-corresponding author on the paper with Siddappa Byrareddy, PhD, professor and vice chair of research in the UNMC Department of Pharmacology and Experimental Neuroscience.
Their work showed completely different mutations in these variants based on regions but also where variants have common lineage. That commonality can be seen between the CAL.20C and B.1.351 variants.
“These variants are continuously evolving and complex in nature, but a proper understanding of what makes each variant unique, its genetic makeup and its interaction with host proteins is critical to developing the most effective vaccine,” Dr. Byrareddy said. “Not only will this help us to design better vaccines now, but greatly help us to be prepared to deal with more mutant variants in the future.”
While related to each other, the variants also developed different mutations.
“B.1.1.7 was more infectious and was able to spread to more people. It slowly dominated and that’s what happened in the United States as B.1.1.7 is now the dominant variant of SARS-CoV-2 in the U.S.,” said Saathvik Kannan, a co-author. Kannan is going to be a 10th grader at Columbia’s Hickman High School this fall but works under Singh at Bond LSC as a computer programmer and researcher.
Once the team identified the related COVID-19 variants, it began to trace their migration in order to see where the mutations were being carried and which regions had a more dense number of infections. The prevalence of variants was key in identifying where they were moving.
The team’s analysis looked at certain mutations in each variant to see how they were related and why some sequences have mutations that others don’t in an attempt to determine how they evolved from their original sequence.
By comparing the structure of each virus, they were able to illustrate how the related mutations moved state by state across the country.
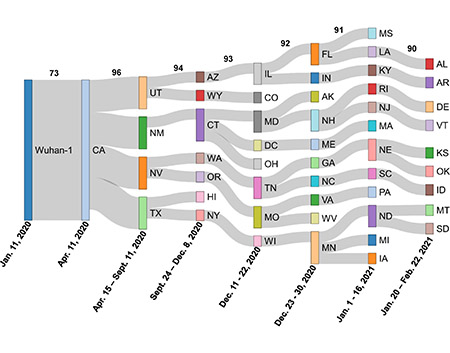 |
This illustration from the paper shows the spread of the variants from California eastward. |
They aligned the first-known CAL.20C sequences from each state and then grouped the sequences based upon the date collected and how closely their structures matched, with cut-offs of 96%, 94%, 93%, 92%, 91% and 90% compared with the previous group.
“It is pretty clear that some states have a higher homology match, which helps us see a pretty linear trend of migration from the Western states to the East in the United States,” said Austin Spratt, a mathematics senior and lead author on the paper. Spratt has worked under Singh at Bond LSC for two years.
The study cautions that the sequences of viruses are being deposited at an unprecedented rate, so it is possible that some conclusions may change as more sequences of variants become available.
Although analysis indicates that mutations only in one variant do not reflect evolution of it, these genetic mistakes may increase infectiousness of a particular variant.
The short period of time, volume of variants and limited travel around the world all point to a greater infectivity than the original (Wuhan-Hu-1) virus. Recent deadly surges in India caused by variant B.1.617.2 or the Delta variant soon will be added to the team’s analysis.

Congrats Siddappa Sir! Nice work.
Congratulations brother 👍👍👍👍🙏🙏🙏🙏