Brahma Lab
The Brahma Lab investigates fundamental mechanisms regulating chromatin structure, how cells use these mechanisms to establish and maintain gene expression programs, and the implications of these mechanisms for human health.
Chromatin is the complex of DNA and histone proteins that organizes our genome into nucleosomes. These nucleosomes regulate access to DNA sequences for regulatory proteins, making chromatin critical in gene expression and other processes. Chromatin-based regulation of gene expression is crucial to the varied functions of cells in our body, and it contributes to epigenetic inheritance. However, defective chromatin regulation can cause developmental disorders and cancer.
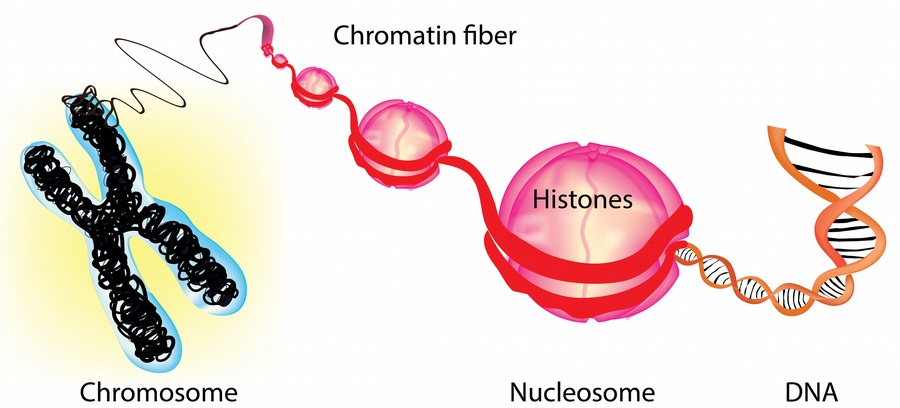
A major focus of our research is on ATP-dependent nucleosome remodeling, which modulates DNA accessibility for transcription, replication, and repair and is crucial for cell-type-specific gene expression during development. Dysfunctions in ATP-dependent nucleosome remodelers are common in human diseases, including more than 25% of all cancers and several developmental disorders. Our lab aims to dissect the biochemical mechanisms of ATP-dependent nucleosome remodeling and understand how it integrates with other chromatin regulators and processes to establish and maintain unique chromatin landscapes that define cellular identity. We take a highly interdisciplinary approach utilizing cutting-edge structural and functional epigenomics, biochemistry, and bioinformatics techniques to achieve our research objectives.
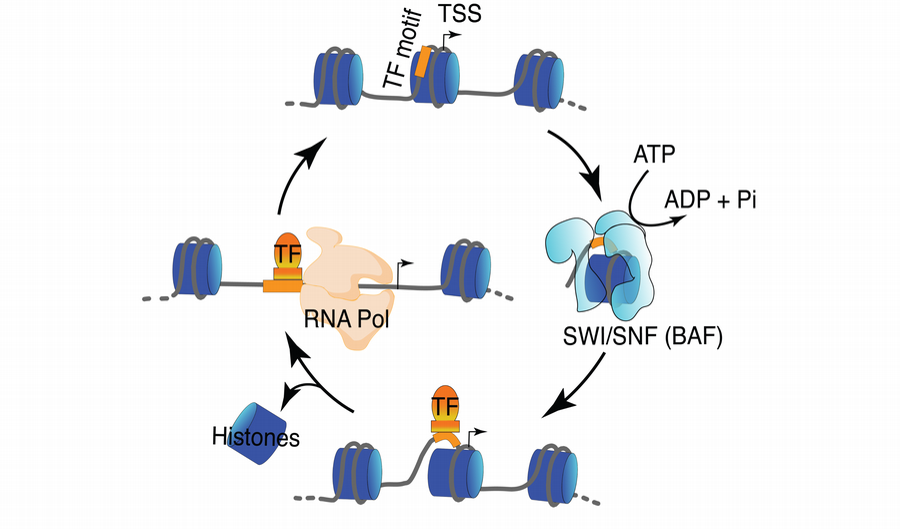
Chromatin structure is dynamic, and ATP-dependent nucleosome remodelers such as SWI/SNF or BAF regulate transcription factor (TF) and RNA Polymerase binding.
Ongoing projects include:
- Investigating the structural and functional aspects of how nucleosome remodelers and gene-regulatory proteins such as transcription factors and RNA Polymerase interact with chromatin.
- Determining how chromatin regulators alter histone-DNA interactions in nucleosomes and modify chromatin structure at high spatial and temporal resolution.
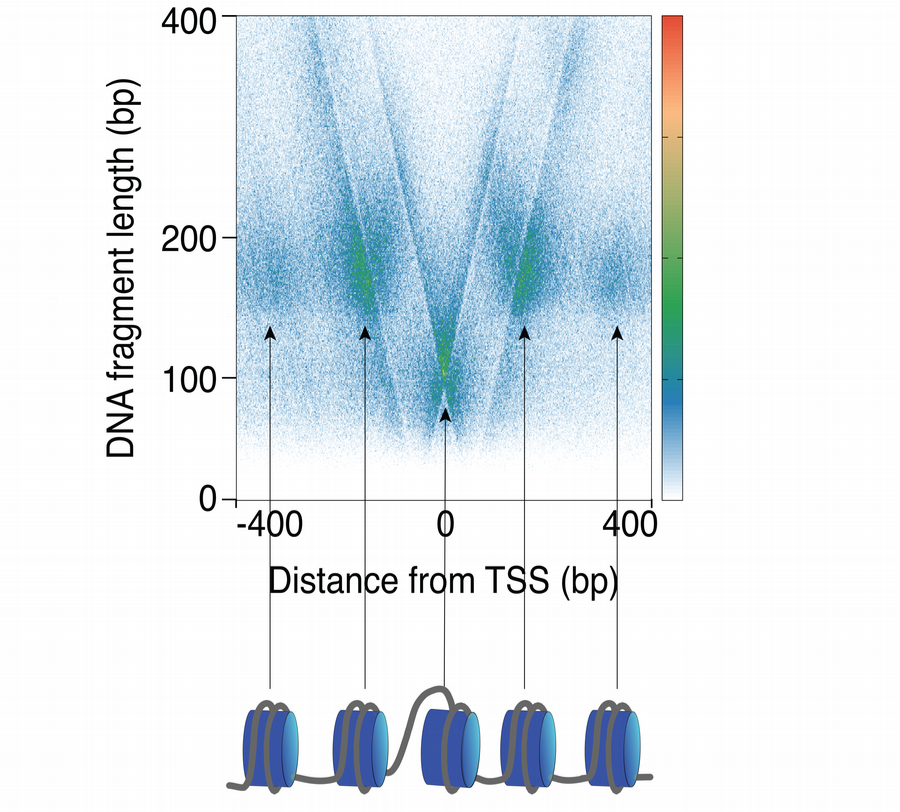
We use techniques to map protein-DNA interaction dynamics at high spatial and temporal resolution genome wide in cells and using purified and reconstituted systems in vitro.
- Dissecting the roles of ATP-dependent nucleosome remodeling in establishing new gene expression programs during cell fate changes, such as embryonic development and cancer.
- We also endeavor to advance the field by developing novel methods for investigating dynamic protein-DNA interactions on chromatin.
Principal Investigator
Sandipan Brahma, PhD
Assistant Professor, UNMC Department of Genetics, Cell Biology and Anatomy
Co-Director, Molecular Genetics & Cell Biology MS Program
